Configuring the pipeline
This feature is in the beta tier. For more information on feature tiers, see API Tiers.
This page explains how to create and configure a link prediction pipeline.
Creating a pipeline
The first step of building a new pipeline is to create one using gds.beta.pipeline.linkPrediction.create.
This stores a trainable pipeline object in the pipeline catalog of type Link prediction training pipeline.
This represents a configurable pipeline that can later be invoked for training, which in turn creates a trained pipeline.
The latter is also a model which is stored in the catalog with type LinkPrediction.
Syntax
CALL gds.beta.pipeline.linkPrediction.create(
pipelineName: String
)
YIELD
name: String,
nodePropertySteps: List of Map,
featureSteps: List of Map,
splitConfig: Map,
autoTuningConfig: Map,
parameterSpace: List of Map| Name | Type | Description |
|---|---|---|
pipelineName |
String |
The name of the created pipeline. |
| Name | Type | Description |
|---|---|---|
name |
String |
Name of the pipeline. |
nodePropertySteps |
List of Map |
List of configurations for node property steps. |
featureSteps |
List of Map |
List of configurations for feature steps. |
splitConfig |
Map |
Configuration to define the split before the model training. |
autoTuningConfig |
Map |
Configuration to define the behavior of auto-tuning. |
parameterSpace |
List of Map |
List of parameter configurations for models which the train mode uses for model selection. |
Example
CALL gds.beta.pipeline.linkPrediction.create('pipe')| name | nodePropertySteps | featureSteps | splitConfig | autoTuningConfig | parameterSpace |
|---|---|---|---|---|---|
"pipe" |
[] |
[] |
{negativeSamplingRatio=1.0, testFraction=0.1, trainFraction=0.1, validationFolds=3} |
{maxTrials=10} |
{LogisticRegression=[], MultilayerPerceptron=[], RandomForest=[]} |
This shows that the newly created pipeline does not contain any steps yet, and has defaults for the split and train parameters.
Adding node properties
A link prediction pipeline can execute one or several GDS algorithms in mutate mode that create node properties in the projected graph. Such steps producing node properties can be chained one after another and created properties can also be used to add features. Moreover, the node property steps that are added to the pipeline will be executed both when training a pipeline and when the trained model is applied for prediction.
The name of the procedure that should be added can be a fully qualified GDS procedure name ending with .mutate.
The ending .mutate may be omitted and one may also use shorthand forms such as node2vec instead of gds.node2vec.mutate.
But please note that a tier qualification must still be given as part of the name.
For example, pre-processing algorithms can be used as node property steps.
Syntax
CALL gds.beta.pipeline.linkPrediction.addNodeProperty(
pipelineName: String,
procedureName: String,
procedureConfiguration: Map
)
YIELD
name: String,
nodePropertySteps: List of Map,
featureSteps: List of Map,
splitConfig: Map,
autoTuningConfig: Map,
parameterSpace: List of Map| Name | Type | Description |
|---|---|---|
pipelineName |
String |
The name of the pipeline. |
procedureName |
String |
The name of the procedure to be added to the pipeline. |
procedureConfiguration |
Map |
The map used to generate the configuration of the procedure. It includes procedure specific configurations except |
| Name | Type | Default | Description |
|---|---|---|---|
contextNodeLabels |
List of String |
|
Additional node labels which are added as context. |
contextRelationshipTypes |
List of String |
|
Additional relationship types which are added as context. |
During training, the context configuration is combined with the train configuration to produce the final node label and relationship type filter for each node property step.
| Name | Type | Description |
|---|---|---|
name |
String |
Name of the pipeline. |
nodePropertySteps |
List of Map |
List of configurations for node property steps. |
featureSteps |
List of Map |
List of configurations for feature steps. |
splitConfig |
Map |
Configuration to define the split before the model training. |
autoTuningConfig |
Map |
Configuration to define the behavior of auto-tuning. |
parameterSpace |
List of Map |
List of parameter configurations for models which the train mode uses for model selection. |
Example
CALL gds.beta.pipeline.linkPrediction.addNodeProperty('pipe', 'fastRP', {
mutateProperty: 'embedding',
embeddingDimension: 256,
randomSeed: 42
})| name | nodePropertySteps | featureSteps | splitConfig | autoTuningConfig | parameterSpace |
|---|---|---|---|---|---|
"pipe" |
[{config={contextNodeLabels=[], contextRelationshipTypes=[], embeddingDimension=256, mutateProperty="embedding", randomSeed=42}, name="gds.fastRP.mutate"}] |
[] |
{negativeSamplingRatio=1.0, testFraction=0.1, trainFraction=0.1, validationFolds=3} |
{maxTrials=10} |
{LogisticRegression=[], MultilayerPerceptron=[], RandomForest=[]} |
The pipeline will now execute the fastRP algorithm in mutate mode both before training a model, and when the trained model is applied for prediction.
This ensures the embedding property can be used as an input for link features.
Adding link features
A Link Prediction pipeline executes a sequence of steps to compute the features used by a machine learning model. A feature step computes a vector of features for given node pairs. For each node pair, the results are concatenated into a single link feature vector. The order of the features in the link feature vector follows the order of the feature steps. Like with node property steps, the feature steps are also executed both at training and prediction time. The supported methods for obtaining features are described below.
Syntax
CALL gds.beta.pipeline.linkPrediction.addFeature(
pipelineName: String,
featureType: String,
configuration: Map
)
YIELD
name: String,
nodePropertySteps: List of Map,
featureSteps: List of Map,
splitConfig: Map,
autoTuningConfig: Map,
parameterSpace: List of Map| Name | Type | Description |
|---|---|---|
pipelineName |
String |
The name of the pipeline. |
featureType |
String |
The featureType determines the method used for computing the link feature. See supported types. |
configuration |
Map |
Configuration for adding the link feature. |
| Name | Type | Default | Description |
|---|---|---|---|
nodeProperties |
List of String |
no |
The names of the node properties that should be used as input. |
| Name | Type | Description |
|---|---|---|
name |
String |
Name of the pipeline. |
nodePropertySteps |
List of Map |
List of configurations for node property steps. |
featureSteps |
List of Map |
List of configurations for feature steps. |
splitConfig |
Map |
Configuration to define the split before the model training. |
autoTuningConfig |
Map |
Configuration to define the behavior of auto-tuning. |
parameterSpace |
List of Map |
List of parameter configurations for models which the train mode uses for model selection. |
Supported feature types
A feature step can use node properties that exist in the input graph or are added by the pipeline.
For each node in each potential link, the values of nodeProperties are concatenated, in the configured order, into a vector f.
That is, for each potential link the feature vector for the source node, , is combined with the one for the target node,
, into a single feature vector f.
The supported types of features can then be described as follows:
| Feature Type | Formula / Description |
|---|---|
L2 |
|
HADAMARD |
|
COSINE |
|
SAME_CATEGORY |
The feature is |
Example
CALL gds.beta.pipeline.linkPrediction.addFeature('pipe', 'hadamard', {
nodeProperties: ['embedding', 'age']
}) YIELD featureSteps| featureSteps |
|---|
[{config={nodeProperties=["embedding", "age"]}, name="HADAMARD"}] |
When executing the pipeline, the nodeProperties must be either present in the input graph, or created by a previous node property step.
For example, the embedding property could be created by the previous example, and we expect age to already be present in the in-memory graph used as input, at train and predict time.
Configuring the relationship splits
Link Prediction training pipelines manage splitting the relationships into several sets and add sampled negative relationships to some of these sets. Configuring the splitting is optional, and if omitted, splitting will be done using default settings.
The splitting configuration of a pipeline can be inspected by using gds.model.list and possibly only yielding splitConfig.
The splitting of relationships proceeds internally in the following steps:
-
The graph is filtered according to specified
sourceNodeLabel,targetNodeLabelandtargetRelationshipType, which are configured at train time. -
The relationships remaining after filtering we call positive, they are split into
test,trainandfeature-inputsets. -
Random negative relationships, which conform to the
sourceNodeLabelandtargetNodeLabelfilter, are added to thetestandtrainsets.-
The number of negative relationships in each set is the number of positive ones multiplied by the
negativeSamplingRatio. -
The negative relationships do not coincide with positive relationships.
-
If
negativeRelationshipTypeis specified, then instead of sampling, all relationships of this type in the graph are partitioned according to thetestandtrainset size ratio and added as negative relationships. All relationships ofnegativeRelationshipTypemust also conform to thesourceNodeLabelandtargetNodeLabelfilter.
-
The positive and negative relationships are given relationship weights of 1.0 and 0.0 respectively so that they can be distinguished.
The train and test relationship sets are used for:
-
determining the label (positive or negative) for each training or test example
-
identifying the node pair for which link features are to be computed
However, they are not used by the algorithms run in the node property steps. The reason for this is that otherwise the model would use the prediction target (existence of a relationship) as a feature.
Each node property step uses a feature-input graph.
The feature-input graph has nodes with sourceNodeLabel, targetNodeLabel and contextNodeLabels and the relationships from the feature-input set plus those of contextRelationshipTypes.
This graph is used for computing node properties and features which depend on node properties.
The node properties generated in the feature-input graph are used in training and testing.
Syntax
CALL gds.beta.pipeline.linkPrediction.configureSplit(
pipelineName: String,
configuration: Map
)
YIELD
name: String,
nodePropertySteps: List of Map,
featureSteps: List of Map,
splitConfig: Map,
autoTuningConfig: Map,
parameterSpace: List of Map| Name | Type | Description |
|---|---|---|
pipelineName |
String |
The name of the pipeline. |
configuration |
Map |
Configuration for splitting the relationships. |
| Name | Type | Default | Description |
|---|---|---|---|
validationFolds |
Integer |
3 |
Number of divisions of the training graph used during model selection. |
testFraction |
Double |
0.1 |
Fraction of the graph reserved for testing. Must be in the range (0, 1). |
trainFraction |
Double |
0.1 |
Fraction of the test-complement set reserved for training. Must be in the range (0, 1). |
negativeSamplingRatio |
Double |
1.0 |
The desired ratio of negative to positive samples in the test and train set. More details here. It is a mutually exclusive parameter with |
negativeRelationshipType |
String |
n/a |
Specifies which relationships should be used as negative relationships, added to the |
| Name | Type | Description |
|---|---|---|
name |
String |
Name of the pipeline. |
nodePropertySteps |
List of Map |
List of configurations for node property steps. |
featureSteps |
List of Map |
List of configurations for feature steps. |
splitConfig |
Map |
Configuration to define the split before the model training. |
autoTuningConfig |
Map |
Configuration to define the behavior of auto-tuning. |
parameterSpace |
List of Map |
List of parameter configurations for models which the train mode uses for model selection. |
Example
CALL gds.beta.pipeline.linkPrediction.configureSplit('pipe', {
testFraction: 0.25,
trainFraction: 0.6,
validationFolds: 3
})
YIELD splitConfig| splitConfig |
|---|
{negativeSamplingRatio=1.0, testFraction=0.25, trainFraction=0.6, validationFolds=3} |
We now reconfigured the splitting of the pipeline, which will be applied during training.
As an example, consider a graph with nodes 'Person' and 'City' and relationships 'KNOWS', 'BORN' and 'LIVES'. Please note that this is the same example as in Training the pipeline.
Suppose we filter by sourceNodeLabel and targetNodeLabel being Person and targetRelationshipType being KNOWS.
The filtered graph looks like the following:

The filtered graph has 12 relationships.
If we configure split with testFraction 0.25 and negativeSamplingRatio 1, it randomly picks 12 * 0.25 = 3 positive relationships plus 1 * 3 = 3 negative relationship as the test set.
Then if trainFraction is 0.6 and negativeSamplingRatio 1, it randomly picks 9 * 0.6 = 5.4 ≈ 5 positive relationships plus 1 * 5 = 5 negative relationship as the train set.
The remaining 12 * (1 - 0.25) * (1 - 0.6) = 3.6 ≈ 4 relationships in yellow is the feature-input set.
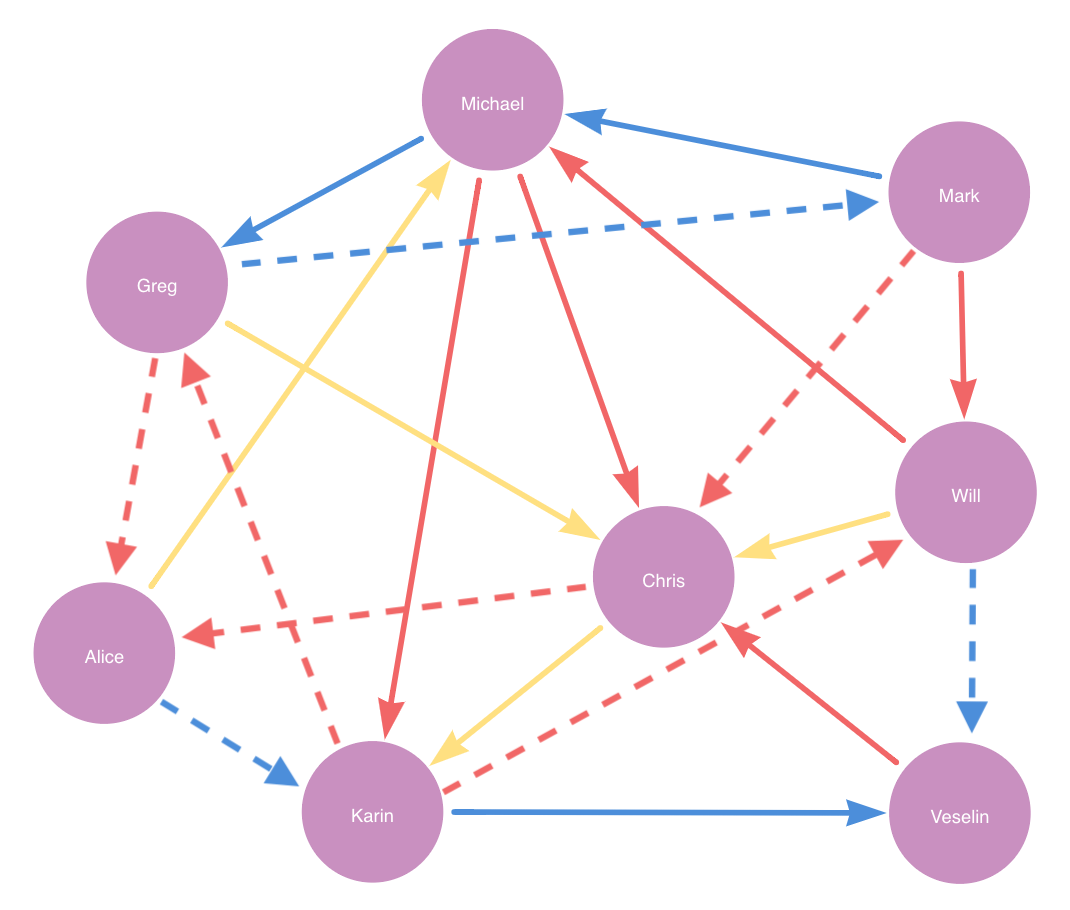
test set is in blue, train set in red and feature-input set in yellow. Dashed lines represent negative relationships.Suppose for example a node property step is added with contextNodeLabel City and contextRelationshipType BORN.
Then the feature-input graph for that step would be:
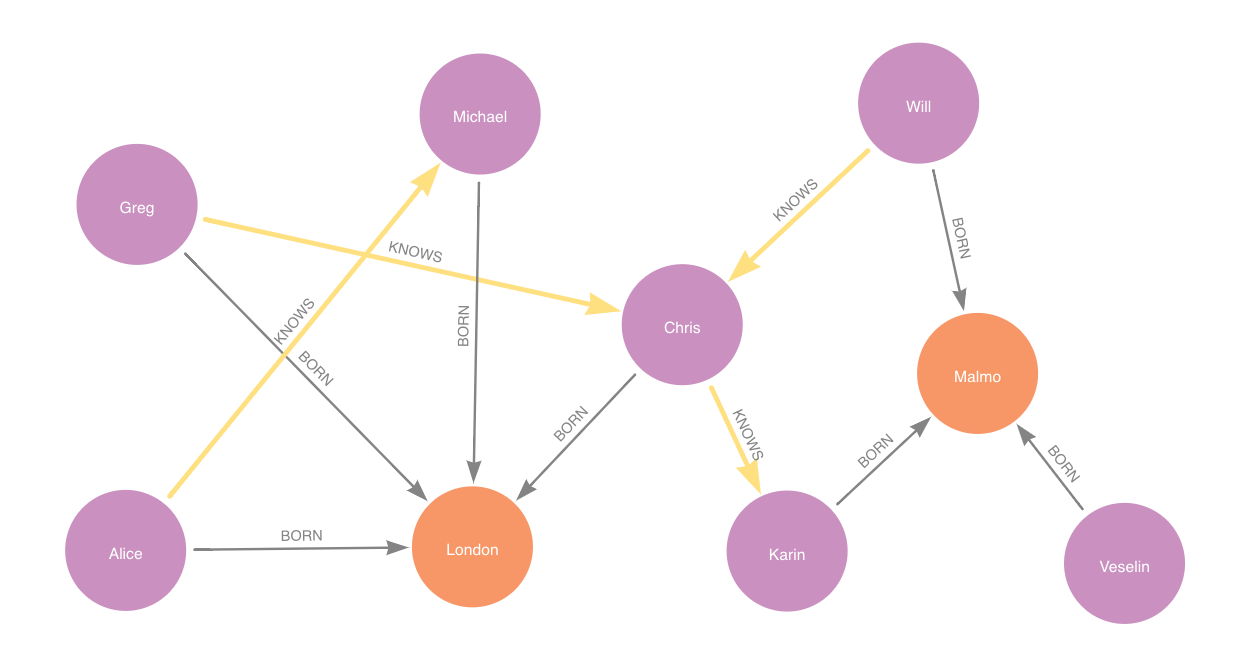
feature-input set is in yellow.Adding model candidates
A pipeline contains a collection of configurations for model candidates which is initially empty. This collection is called the parameter space. Each model candidate configuration contains either fixed values or ranges for training parameters. When a range is present, values from the range are determined automatically by an auto-tuning algorithm, see Auto-tuning. One or more model configurations must be added to the parameter space of the training pipeline, using one of the following procedures:
-
gds.beta.pipeline.linkPrediction.addLogisticRegression -
gds.beta.pipeline.linkPrediction.addRandomForest -
gds.alpha.pipeline.linkPrediction.addMLP
For information about the available training methods in GDS, logistic regression, random forest and multilayer perceptron, see Training methods.
In Training the pipeline, we explain further how the configured model candidates are trained, evaluated and compared.
The parameter space of a pipeline can be inspected using gds.model.list and optionally yielding only parameterSpace.
|
At least one model candidate must be added to the pipeline before training it. |
Syntax
CALL gds.beta.pipeline.linkPrediction.addLogisticRegression(
pipelineName: String,
config: Map
)
YIELD
name: String,
nodePropertySteps: List of Map,
featureSteps: List of Map,
splitConfig: Map,
autoTuningConfig: Map,
parameterSpace: Map| Name | Type | Description |
|---|---|---|
pipelineName |
String |
The name of the pipeline. |
config |
Map |
The logistic regression config for a model candidate. The allowed parameters for a model are defined in the next table. |
| Name | Type | Default | Optional | Description |
|---|---|---|---|---|
|
Integer or Map [1] |
|
yes |
Number of nodes per batch. |
|
Integer or Map [1] |
|
yes |
Minimum number of training epochs. |
|
Integer or Map [1] |
|
yes |
Maximum number of training epochs. |
|
Float or Map [1] |
|
yes |
The learning rate determines the step size at each epoch while moving in the direction dictated by the Adam optimizer for minimizing the loss. |
|
Integer or Map [1] |
|
yes |
Maximum number of unproductive consecutive epochs. |
|
Float or Map [1] |
|
yes |
The minimal improvement of the loss to be considered productive. |
|
Float or Map [1] |
|
yes |
Penalty used for the logistic regression. By default, no penalty is applied. |
|
Float or Map [1] |
|
yes |
Exponent for the focal loss factor, to make the model focus more on hard, misclassified examples in the train set. The default of |
|
List of Float |
|
yes |
Weights for each class in loss function. The list must have length 2. The first weight is for negative examples (missing relationships), and the second for positive examples (actual relationships). |
1. A map should be of the form 2. Ranges for this parameter are auto-tuned on a logarithmic scale. |
||||
| Name | Type | Description |
|---|---|---|
name |
String |
Name of the pipeline. |
nodePropertySteps |
List of Map |
List of configurations for node property steps. |
featureSteps |
List of Map |
List of configurations for feature steps. |
splitConfig |
Map |
Configuration to define the split before the model training. |
autoTuningConfig |
Map |
Configuration to define the behavior of auto-tuning. |
parameterSpace |
List of Map |
List of parameter configurations for models which the train mode uses for model selection. |
CALL gds.beta.pipeline.linkPrediction.addRandomForest(
pipelineName: String,
config: Map
)
YIELD
name: String,
nodePropertySteps: List of Map,
featureSteps: List of Map,
splitConfig: Map,
autoTuningConfig: Map,
parameterSpace: Map| Name | Type | Description |
|---|---|---|
pipelineName |
String |
The name of the pipeline. |
config |
Map |
The random forest config for a model candidate. The allowed parameters for a model are defined in the next table. |
| Name | Type | Default | Optional | Description |
|---|---|---|---|---|
maxFeaturesRatio |
Float or Map [3] |
|
yes |
The ratio of features to consider when looking for the best split |
numberOfSamplesRatio |
Float or Map [3] |
|
yes |
The ratio of samples to consider per decision tree. We use sampling with replacement. A value of |
numberOfDecisionTrees |
Integer or Map [3] |
|
yes |
The number of decision trees. |
maxDepth |
Integer or Map [3] |
|
yes |
The maximum depth of a decision tree. |
minLeafSize |
Integer or Map [3] |
|
yes |
The minimum number of samples for a leaf node in a decision tree. Must be strictly smaller than |
minSplitSize |
Integer or Map [3] |
|
yes |
The minimum number of samples required to split an internal node in a decision tree. Must be strictly larger than |
criterion |
String |
|
yes |
The impurity criterion used to evaluate potential node splits during decision tree training. Valid options are |
3. A map should be of the form |
||||
| Name | Type | Description |
|---|---|---|
name |
String |
Name of the pipeline. |
nodePropertySteps |
List of Map |
List of configurations for node property steps. |
featureSteps |
List of Map |
List of configurations for feature steps. |
splitConfig |
Map |
Configuration to define the split before the model training. |
autoTuningConfig |
Map |
Configuration to define the behavior of auto-tuning. |
parameterSpace |
List of Map |
List of parameter configurations for models which the train mode uses for model selection. |
CALL gds.alpha.pipeline.linkPrediction.addMLP(
pipelineName: String,
config: Map
)
YIELD
name: String,
nodePropertySteps: List of Map,
featureSteps: List of Map,
splitConfig: Map,
autoTuningConfig: Map,
parameterSpace: Map| Name | Type | Description |
|---|---|---|
pipelineName |
String |
The name of the pipeline. |
config |
Map |
The multilayer perceptron config for a model candidate. The allowed parameters for a model are defined in the next table. |
| Name | Type | Default | Optional | Description |
|---|---|---|---|---|
batchSize |
Integer or Map [4] |
|
yes |
Number of nodes per batch. |
minEpochs |
Integer or Map [4] |
|
yes |
Minimum number of training epochs. |
maxEpochs |
Integer or Map [4] |
|
yes |
Maximum number of training epochs. |
learningRate [5] |
Float or Map [4] |
|
yes |
The learning rate determines the step size at each epoch while moving in the direction dictated by the Adam optimizer for minimizing the loss. |
patience |
Integer or Map [4] |
|
yes |
Maximum number of unproductive consecutive epochs. |
tolerance [5] |
Float or Map [4] |
|
yes |
The minimal improvement of the loss to be considered productive. |
penalty [5] |
Float or Map [4] |
|
yes |
Penalty used for the logistic regression. By default, no penalty is applied. |
hiddenLayerSizes |
List of Integers |
|
yes |
List of integers representing number of neurons in each layer. The default value specifies an MLP with 1 hidden layer of 100 neurons. |
focusWeight |
Float or Map [4] |
|
yes |
Exponent for the focal loss factor, to make the model focus more on hard, misclassified examples in the train set. The default of |
classWeights |
List of Float |
|
yes |
Weights for each class in cross-entropy loss. The list must have length 2. The first weight is for negative examples (missing relationships), and the second for positive examples (actual relationships). |
4. A map should be of the form 5. Ranges for this parameter are auto-tuned on a logarithmic scale. |
||||
| Name | Type | Description |
|---|---|---|
name |
String |
Name of the pipeline. |
nodePropertySteps |
List of Map |
List of configurations for node property steps. |
featureSteps |
List of Map |
List of configurations for feature steps. |
splitConfig |
Map |
Configuration to define the split before the model training. |
autoTuningConfig |
Map |
Configuration to define the behavior of auto-tuning. |
parameterSpace |
List of Map |
List of parameter configurations for models which the train mode uses for model selection. |
Example
We can add multiple model candidates to our pipeline.
CALL gds.beta.pipeline.linkPrediction.addLogisticRegression('pipe')
YIELD parameterSpaceCALL gds.beta.pipeline.linkPrediction.addRandomForest('pipe', {numberOfDecisionTrees: 10})
YIELD parameterSpaceCALL gds.alpha.pipeline.linkPrediction.addMLP('pipe',
{hiddenLayerSizes: [4, 2], penalty: 0.5, patience: 2, classWeights: [0.55, 0.45], focusWeight: {range: [0.0, 0.1]}})
YIELD parameterSpaceCALL gds.beta.pipeline.linkPrediction.addLogisticRegression('pipe', {maxEpochs: 500, penalty: {range: [1e-4, 1e2]}})
YIELD parameterSpace
RETURN parameterSpace.RandomForest AS randomForestSpace, parameterSpace.LogisticRegression AS logisticRegressionSpace, parameterSpace.MultilayerPerceptron AS MultilayerPerceptronSpace| randomForestSpace | logisticRegressionSpace | MultilayerPerceptronSpace |
|---|---|---|
[{criterion="GINI", maxDepth=2147483647, methodName="RandomForest", minLeafSize=1, minSplitSize=2, numberOfDecisionTrees=10, numberOfSamplesRatio=1.0}] |
[{batchSize=100, classWeights=[], focusWeight=0.0, learningRate=0.001, maxEpochs=100, methodName="LogisticRegression", minEpochs=1, patience=1, penalty=0.0, tolerance=0.001}, {batchSize=100, classWeights=[], focusWeight=0.0, learningRate=0.001, maxEpochs=500, methodName="LogisticRegression", minEpochs=1, patience=1, penalty={range=[0.0001, 100.0]}, tolerance=0.001}] |
[{batchSize=100, classWeights=[0.55, 0.45], focusWeight={range=[0.0, 0.1]}, hiddenLayerSizes=[4, 2], learningRate=0.001, maxEpochs=100, methodName="MultilayerPerceptron", minEpochs=1, patience=2, penalty=0.5, tolerance=0.001}] |
The parameterSpace in the pipeline now contains the four different model candidates, expanded with the default values.
Each specified model candidate will be tried out during the model selection in training.
|
These are somewhat naive examples of how to add and configure model candidates. Please see Training methods for more information on how to tune the configuration parameters of each method. |
Configuring Auto-tuning
In order to find good models, the pipeline supports automatically tuning the parameters of the training algorithm. Optionally, the procedure described below can be used to configure the auto-tuning behavior. Otherwise, default auto-tuning configuration is used. Currently, it is only possible to configure the maximum number trials of hyper-parameter settings which are evaluated.
Syntax
CALL gds.alpha.pipeline.linkPrediction.configureAutoTuning(
pipelineName: String,
configuration: Map
)
YIELD
name: String,
nodePropertySteps: List of Map,
featureSteps: List of Map,
splitConfig: Map,
autoTuningConfig: Map,
parameterSpace: List of Map| Name | Type | Description |
|---|---|---|
pipelineName |
String |
The name of the created pipeline. |
configuration |
Map |
The configuration for auto-tuning. |
| Name | Type | Default | Description |
|---|---|---|---|
maxTrials |
Integer |
10 |
The value of |
| Name | Type | Description |
|---|---|---|
name |
String |
Name of the pipeline. |
nodePropertySteps |
List of Map |
List of configurations for node property steps. |
featureSteps |
List of Map |
List of configurations for feature steps. |
splitConfig |
Map |
Configuration to define the split before the model training. |
autoTuningConfig |
Map |
Configuration to define the behavior of auto-tuning. |
parameterSpace |
List of Map |
List of parameter configurations for models which the train mode uses for model selection. |
Example
CALL gds.alpha.pipeline.linkPrediction.configureAutoTuning('pipe', {
maxTrials: 2
}) YIELD autoTuningConfig| autoTuningConfig |
|---|
{maxTrials=2} |
We now reconfigured the auto-tuning to try out at most 2 model candidates during training.